Zero Shot Segmentation medical imaging
Papers and Code
MRAD: Zero-Shot Anomaly Detection with Memory-Driven Retrieval
Jan 31, 2026Zero-shot anomaly detection (ZSAD) often leverages pretrained vision or vision-language models, but many existing methods use prompt learning or complex modeling to fit the data distribution, resulting in high training or inference cost and limited cross-domain stability. To address these limitations, we propose Memory-Retrieval Anomaly Detection method (MRAD), a unified framework that replaces parametric fitting with a direct memory retrieval. The train-free base model, MRAD-TF, freezes the CLIP image encoder and constructs a two-level memory bank (image-level and pixel-level) from auxiliary data, where feature-label pairs are explicitly stored as keys and values. During inference, anomaly scores are obtained directly by similarity retrieval over the memory bank. Based on the MRAD-TF, we further propose two lightweight variants as enhancements: (i) MRAD-FT fine-tunes the retrieval metric with two linear layers to enhance the discriminability between normal and anomaly; (ii) MRAD-CLIP injects the normal and anomalous region priors from the MRAD-FT as dynamic biases into CLIP's learnable text prompts, strengthening generalization to unseen categories. Across 16 industrial and medical datasets, the MRAD framework consistently demonstrates superior performance in anomaly classification and segmentation, under both train-free and training-based settings. Our work shows that fully leveraging the empirical distribution of raw data, rather than relying only on model fitting, can achieve stronger anomaly detection performance. The code will be publicly released at https://github.com/CROVO1026/MRAD.
Opportunistic Promptable Segmentation: Leveraging Routine Radiological Annotations to Guide 3D CT Lesion Segmentation
Jan 30, 2026The development of machine learning models for CT imaging depends on the availability of large, high-quality, and diverse annotated datasets. Although large volumes of CT images and reports are readily available in clinical picture archiving and communication systems (PACS), 3D segmentations of critical findings are costly to obtain, typically requiring extensive manual annotation by radiologists. On the other hand, it is common for radiologists to provide limited annotations of findings during routine reads, such as line measurements and arrows, that are often stored in PACS as GSPS objects. We posit that these sparse annotations can be extracted along with CT volumes and converted into 3D segmentations using promptable segmentation models, a paradigm we term Opportunistic Promptable Segmentation. To enable this paradigm, we propose SAM2CT, the first promptable segmentation model designed to convert radiologist annotations into 3D segmentations in CT volumes. SAM2CT builds upon SAM2 by extending the prompt encoder to support arrow and line inputs and by introducing Memory-Conditioned Memories (MCM), a memory encoding strategy tailored to 3D medical volumes. On public lesion segmentation benchmarks, SAM2CT outperforms existing promptable segmentation models and similarly trained baselines, achieving Dice similarity coefficients of 0.649 for arrow prompts and 0.757 for line prompts. Applying the model to pre-existing GSPS annotations from a clinical PACS (N = 60), SAM2CT generates 3D segmentations that are clinically acceptable or require only minor adjustments in 87% of cases, as scored by radiologists. Additionally, SAM2CT demonstrates strong zero-shot performance on select Emergency Department findings. These results suggest that large-scale mining of historical GSPS annotations represents a promising and scalable approach for generating 3D CT segmentation datasets.
Synthetic Volumetric Data Generation Enables Zero-Shot Generalization of Foundation Models in 3D Medical Image Segmentation
Jan 18, 2026Foundation models such as Segment Anything Model 2 (SAM 2) exhibit strong generalization on natural images and videos but perform poorly on medical data due to differences in appearance statistics, imaging physics, and three-dimensional structure. To address this gap, we introduce SynthFM-3D, an analytical framework that mathematically models 3D variability in anatomy, contrast, boundary definition, and noise to generate synthetic data for training promptable segmentation models without real annotations. We fine-tuned SAM 2 on 10,000 SynthFM-3D volumes and evaluated it on eleven anatomical structures across three medical imaging modalities (CT, MR, ultrasound) from five public datasets. SynthFM-3D training led to consistent and statistically significant Dice score improvements over the pretrained SAM 2 baseline, demonstrating stronger zero-shot generalization across modalities. When compared with the supervised SAM-Med3D model on unseen cardiac ultrasound data, SynthFM-3D achieved 2-3x higher Dice scores, establishing analytical 3D data modeling as an effective pathway to modality-agnostic medical segmentation.
EchoVLM: Measurement-Grounded Multimodal Learning for Echocardiography
Dec 13, 2025Echocardiography is the most widely used imaging modality in cardiology, yet its interpretation remains labor-intensive and inherently multimodal, requiring view recognition, quantitative measurements, qualitative assessments, and guideline-based reasoning. While recent vision-language models (VLMs) have achieved broad success in natural images and certain medical domains, their potential in echocardiography has been limited by the lack of large-scale, clinically grounded image-text datasets and the absence of measurement-based reasoning central to echo interpretation. We introduce EchoGround-MIMIC, the first measurement-grounded multimodal echocardiography dataset, comprising 19,065 image-text pairs from 1,572 patients with standardized views, structured measurements, measurement-grounded captions, and guideline-derived disease labels. Building on this resource, we propose EchoVLM, a vision-language model that incorporates two novel pretraining objectives: (i) a view-informed contrastive loss that encodes the view-dependent structure of echocardiographic imaging, and (ii) a negation-aware contrastive loss that distinguishes clinically critical negative from positive findings. Across five types of clinical applications with 36 tasks spanning multimodal disease classification, image-text retrieval, view classification, chamber segmentation, and landmark detection, EchoVLM achieves state-of-the-art performance (86.5% AUC in zero-shot disease classification and 95.1% accuracy in view classification). We demonstrate that clinically grounded multimodal pretraining yields transferable visual representations and establish EchoVLM as a foundation model for end-to-end echocardiography interpretation. We will release EchoGround-MIMIC and the data curation code, enabling reproducibility and further research in multimodal echocardiography interpretation.
VoxTell: Free-Text Promptable Universal 3D Medical Image Segmentation
Nov 14, 2025We introduce VoxTell, a vision-language model for text-prompted volumetric medical image segmentation. It maps free-form descriptions, from single words to full clinical sentences, to 3D masks. Trained on 62K+ CT, MRI, and PET volumes spanning over 1K anatomical and pathological classes, VoxTell uses multi-stage vision-language fusion across decoder layers to align textual and visual features at multiple scales. It achieves state-of-the-art zero-shot performance across modalities on unseen datasets, excelling on familiar concepts while generalizing to related unseen classes. Extensive experiments further demonstrate strong cross-modality transfer, robustness to linguistic variations and clinical language, as well as accurate instance-specific segmentation from real-world text. Code is available at: https://www.github.com/MIC-DKFZ/VoxTell
Towards Better Ultrasound Video Segmentation Foundation Model: An Empirical study on SAM2 Finetuning from Data Perspective
Nov 07, 2025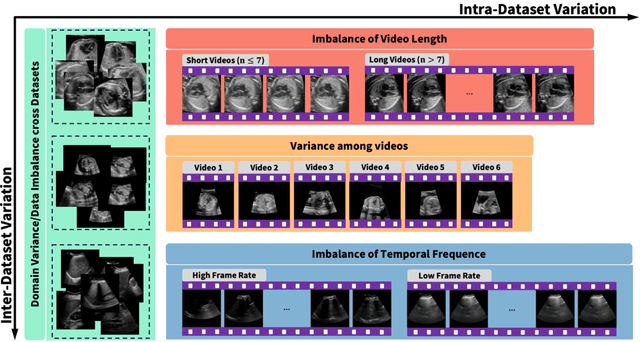

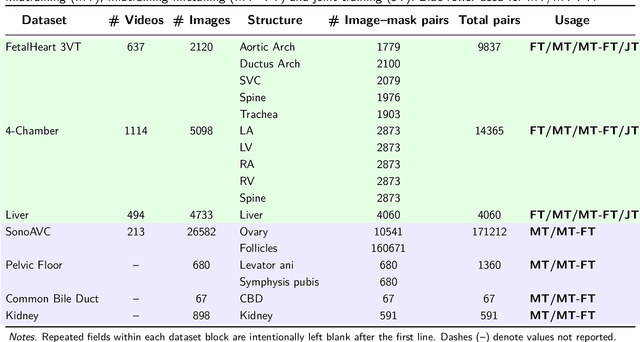
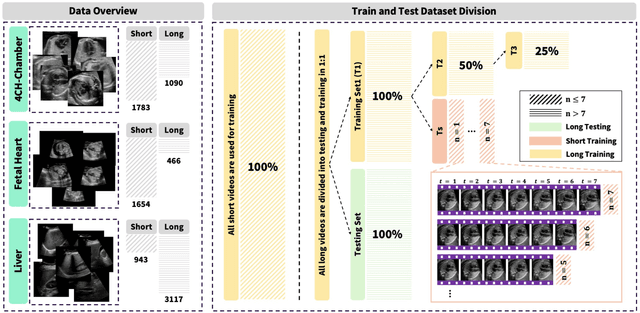
Ultrasound (US) video segmentation remains a challenging problem due to strong inter- and intra-dataset variability, motion artifacts, and limited annotated data. Although foundation models such as Segment Anything Model 2 (SAM2) demonstrate strong zero-shot and prompt-guided segmentation capabilities, their performance deteriorates substantially when transferred to medical imaging domains. Current adaptation studies mainly emphasize architectural modifications, while the influence of data characteristics and training regimes has not been systematically examined. In this study, we present a comprehensive, data-centric investigation of SAM2 adaptation for ultrasound video segmentation. We analyze how training-set size, video duration, and augmentation schemes affect adaptation performance under three paradigms: task-specific fine-tuning, intermediate adaptation, and multi-task joint training, across five SAM2 variants and multiple prompting modes. We further design six ultrasound-specific augmentations, assessing their effect relative to generic strategies. Experiments on three representative ultrasound datasets reveal that data scale and temporal context play a more decisive role than model architecture or initialization. Moreover, joint training offers an efficient compromise between modality alignment and task specialization. This work aims to provide empirical insights for developing efficient, data-aware adaptation pipelines for SAM2 in ultrasound video analysis.
Anatomy-VLM: A Fine-grained Vision-Language Model for Medical Interpretation
Nov 11, 2025Accurate disease interpretation from radiology remains challenging due to imaging heterogeneity. Achieving expert-level diagnostic decisions requires integration of subtle image features with clinical knowledge. Yet major vision-language models (VLMs) treat images as holistic entities and overlook fine-grained image details that are vital for disease diagnosis. Clinicians analyze images by utilizing their prior medical knowledge and identify anatomical structures as important region of interests (ROIs). Inspired from this human-centric workflow, we introduce Anatomy-VLM, a fine-grained, vision-language model that incorporates multi-scale information. First, we design a model encoder to localize key anatomical features from entire medical images. Second, these regions are enriched with structured knowledge for contextually-aware interpretation. Finally, the model encoder aligns multi-scale medical information to generate clinically-interpretable disease prediction. Anatomy-VLM achieves outstanding performance on both in- and out-of-distribution datasets. We also validate the performance of Anatomy-VLM on downstream image segmentation tasks, suggesting that its fine-grained alignment captures anatomical and pathology-related knowledge. Furthermore, the Anatomy-VLM's encoder facilitates zero-shot anatomy-wise interpretation, providing its strong expert-level clinical interpretation capabilities.
CrossMed: A Multimodal Cross-Task Benchmark for Compositional Generalization in Medical Imaging
Nov 14, 2025



Recent advances in multimodal large language models have enabled unified processing of visual and textual inputs, offering promising applications in general-purpose medical AI. However, their ability to generalize compositionally across unseen combinations of imaging modality, anatomy, and task type remains underexplored. We introduce CrossMed, a benchmark designed to evaluate compositional generalization (CG) in medical multimodal LLMs using a structured Modality-Anatomy-Task (MAT) schema. CrossMed reformulates four public datasets, CheXpert (X-ray classification), SIIM-ACR (X-ray segmentation), BraTS 2020 (MRI classification and segmentation), and MosMedData (CT classification) into a unified visual question answering (VQA) format, resulting in 20,200 multiple-choice QA instances. We evaluate two open-source multimodal LLMs, LLaVA-Vicuna-7B and Qwen2-VL-7B, on both Related and Unrelated MAT splits, as well as a zero-overlap setting where test triplets share no Modality, Anatomy, or Task with the training data. Models trained on Related splits achieve 83.2 percent classification accuracy and 0.75 segmentation cIoU, while performance drops significantly under Unrelated and zero-overlap conditions, demonstrating the benchmark difficulty. We also show cross-task transfer, where segmentation performance improves by 7 percent cIoU even when trained using classification-only data. Traditional models (ResNet-50 and U-Net) show modest gains, confirming the broad utility of the MAT framework, while multimodal LLMs uniquely excel at compositional generalization. CrossMed provides a rigorous testbed for evaluating zero-shot, cross-task, and modality-agnostic generalization in medical vision-language models.
VessShape: Few-shot 2D blood vessel segmentation by leveraging shape priors from synthetic images
Oct 31, 2025Semantic segmentation of blood vessels is an important task in medical image analysis, but its progress is often hindered by the scarcity of large annotated datasets and the poor generalization of models across different imaging modalities. A key aspect is the tendency of Convolutional Neural Networks (CNNs) to learn texture-based features, which limits their performance when applied to new domains with different visual characteristics. We hypothesize that leveraging geometric priors of vessel shapes, such as their tubular and branching nature, can lead to more robust and data-efficient models. To investigate this, we introduce VessShape, a methodology for generating large-scale 2D synthetic datasets designed to instill a shape bias in segmentation models. VessShape images contain procedurally generated tubular geometries combined with a wide variety of foreground and background textures, encouraging models to learn shape cues rather than textures. We demonstrate that a model pre-trained on VessShape images achieves strong few-shot segmentation performance on two real-world datasets from different domains, requiring only four to ten samples for fine-tuning. Furthermore, the model exhibits notable zero-shot capabilities, effectively segmenting vessels in unseen domains without any target-specific training. Our results indicate that pre-training with a strong shape bias can be an effective strategy to overcome data scarcity and improve model generalization in blood vessel segmentation.
U-Bench: A Comprehensive Understanding of U-Net through 100-Variant Benchmarking
Oct 08, 2025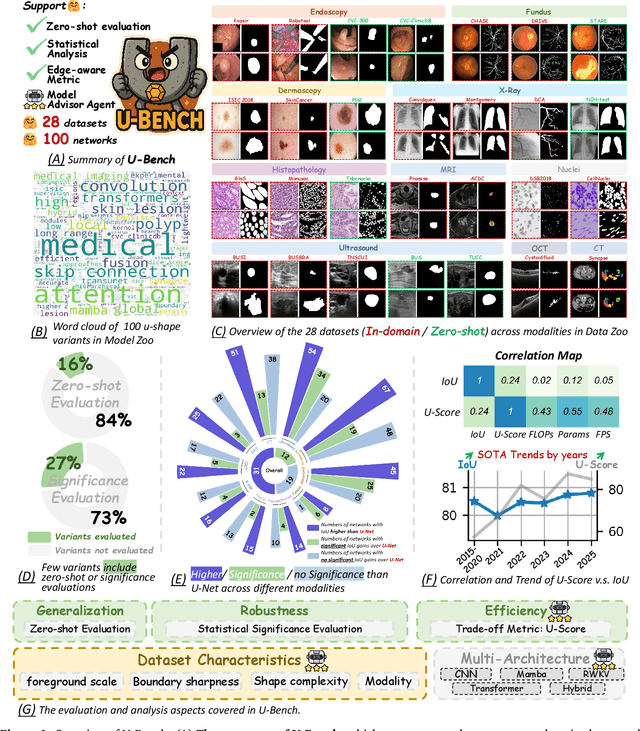


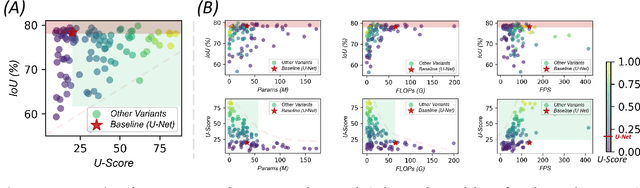
Over the past decade, U-Net has been the dominant architecture in medical image segmentation, leading to the development of thousands of U-shaped variants. Despite its widespread adoption, there is still no comprehensive benchmark to systematically evaluate their performance and utility, largely because of insufficient statistical validation and limited consideration of efficiency and generalization across diverse datasets. To bridge this gap, we present U-Bench, the first large-scale, statistically rigorous benchmark that evaluates 100 U-Net variants across 28 datasets and 10 imaging modalities. Our contributions are threefold: (1) Comprehensive Evaluation: U-Bench evaluates models along three key dimensions: statistical robustness, zero-shot generalization, and computational efficiency. We introduce a novel metric, U-Score, which jointly captures the performance-efficiency trade-off, offering a deployment-oriented perspective on model progress. (2) Systematic Analysis and Model Selection Guidance: We summarize key findings from the large-scale evaluation and systematically analyze the impact of dataset characteristics and architectural paradigms on model performance. Based on these insights, we propose a model advisor agent to guide researchers in selecting the most suitable models for specific datasets and tasks. (3) Public Availability: We provide all code, models, protocols, and weights, enabling the community to reproduce our results and extend the benchmark with future methods. In summary, U-Bench not only exposes gaps in previous evaluations but also establishes a foundation for fair, reproducible, and practically relevant benchmarking in the next decade of U-Net-based segmentation models. The project can be accessed at: https://fenghetan9.github.io/ubench. Code is available at: https://github.com/FengheTan9/U-Bench.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge